Gremlin-Python: Algorithm Development from the Ground Up
We've been working with Gremlin for the past 18 months and simply love it! To share our enthusiasm with the world, we compiled a list of helpful tips that will help you embark on your own GraphDB journey (you’re also more than welcome to join ours 😉. Yes, we are hiring!).

A lot was written about Gremlin, but not as much about the Gremlin-Python variance. Understanding the key differences between the two will get you going (official resources for Gremlin-Python: https://tinkerpop.apache.org/docs/current/reference/#gremlin-python).
The post covers the use of the Gremlin-Python language variance with Neptune as the graph database. We go from the very basics of Gremlin-Python to full-scale optimized algorithm development. It’s a hands-on guide and all of the information is available in the accompanying notebooks. We strongly advise you to give it a go!
Before Getting Started
Run a Gremlin server on your machine:

To play around with the toy graph we build throughout this post, set a local Gremlin server using the following command, on a different shell:
docker run --rm -it -p 8182:8182 --name gremlin tinkerpop/gremlin-serverSet up the Gremlin connection:
Set up your connection by using one of the following methods:
Method 1 - python
After you set up the local gremlin server, you can connect to it with the following code:
import nest_asyncio from gremlin_python.driver.driver_remote_connection
import DriverRemoteConnection
from gremlin_python.process.anonymous_traversal import traversal
nest_asyncio.apply() # to support gremlin event loop in jupyter lab
# set the graph traversal from the local machine:
connection = DriverRemoteConnection("ws://localhost:8182/gremlin", "g") # Connect it to your local server
g = traversal().withRemote(connection)
g.V().count().next() # to validate connection, should return ‘0’Method 2
Install Java.
# Using apt
sudo apt install openjdk-8-jdk
# or using brew
brew install openjdkDownload the Gremlin server’s binary. Optionally, change the configuration. You may, for example, change “id management” conf/tinkergraph-empty.properties to use ANY IdManagers instead of LONG. Run: bin/gremlin-server.sh.
Having Import Issues?
If you run samples of other languages, you will usually come across importing errors. Use the following to avoid these errors:
from gremlin_python.driver.driver_remote_connection import DriverRemoteConnection
from gremlin_python.process.graph_traversal import GraphTraversalSource, __
from gremlin_python.process.traversal import Barrier, Bindings, Cardinality, Column, Direction, Operator, Order, P, Pop, Scope, T, WithOptionsYou can also see the names associated with each library:
print("T: " + str(list(filter(lambda x: not str(x).startswith("_"), dir(T)))))
print("Order: " + str(list(filter(lambda x: not str(x).startswith("_"), dir(Order)))))
print("Cardinality: " + str(list(filter(lambda x: not str(x).startswith("_"), dir(Cardinality)))))
print("Column: " + str(list(filter(lambda x: not str(x).startswith("_"), dir(Column)))))
print("Direction: " + str(list(filter(lambda x: not str(x).startswith("_"), dir(Direction)))))
print("Operator: " + str(list(filter(lambda x: not str(x).startswith("_"), dir(Operator)))))
print("P: " + str(list(filter(lambda x: not str(x).startswith("_"), dir(P)))))
print("Pop: " + str(list(filter(lambda x: not str(x).startswith("_"), dir(Pop)))))
print("Scope: " + str(list(filter(lambda x: not str(x).startswith("_"), dir(Scope)))))
print("Barrier: " + str(list(filter(lambda x: not str(x).startswith("_"), dir(Barrier)))))
print("Bindings: " + str(list(filter(lambda x: not str(x).startswith("_"), dir(Bindings)))))
print("WithOptions: " + str(list(filter(lambda x: not str(x).startswith("_"), dir(WithOptions)))))Traversal Submitting
As opposed to other language variants, in Gremlin-Python, you cannot use a traversal without specifying how to return the results. The common options are: to_list(), to_set(), next(), and iterate().
This is how we use traversal submitting in our toy graph:
Set a Toy Graph
Some basic functions (not optimized) to add vertices and edges:
from typing import Optional
def add_edge(g: GraphTraversalSource, from_id: int, to_id: int, edge_label: str, param: Optional[str] = None):
g.V(from_id).addE(edge_label).to(__.V(to_id)).property("param", param).next()
def add_vertex(g: GraphTraversalSource, vertex_label: str, vertex_id: int, name: Optional[str] = None):
g.addV(vertex_label).property(T.id, vertex_id).property("name", name).next()
def init_toy_graph(g: GraphTraversalSource):
g.V().drop().iterate() # so you can run this cell more than ones
g.E().drop().iterate() # so you can run this cell more than ones
add_vertex(g, "user", 1, name="Olivia")
add_vertex(g, "user", 2, name="Emma")
add_vertex(g, "file", 3, name="your_new_idea.pdf")
add_vertex(g, "file", 4, name="salary.pdf")
add_vertex(g, "file", 5, name="demo.py")
add_vertex(g, "file", 6, name="blog.html")
add_vertex(g, "drive", 7, name="my_drive")
add_edge(g, 1, 2, "edit")
add_edge(g, 1, 3, "edit")
add_edge(g, 1, 4, "view")
add_edge(g, 2, 5, "print")
add_edge(g, 2, 6, "edit")
add_edge(g, 3, 7, "located_in")
init_toy_graph(g)
# count how many
print(g.V().valueMap(True).toList()) # get a list of all of the vertices
"""
[{<T.id: 1>: 1, <T.label: 4>: 'user', 'name': ['Olivia']}, {<T.id: 1>: 2, <T.label: 4>: 'user', 'name': ['Emma']}, {<T.id: 1>: 3, <T.label: 4>: 'file', 'name': ['your_new_idea.pdf']}, {<T.id: 1>: 4, <T.label: 4>: 'file', 'name': ['salary.pdf']}, {<T.id: 1>: 5, <T.label: 4>: 'file', 'name': ['demo.py']}, {<T.id: 1>: 6, <T.label: 4>: 'file', 'name': ['blog.html']}, {<T.id: 1>: 7, <T.label: 4>: 'drive', 'name': ['my_drive']}]
"""
print(g.E().valueMap(True).toList()) # get a list of all of the edges
"""
[{<T.id: 1>: 1024, <T.label: 4>: 'view'}, {<T.id: 1>: 1025, <T.label: 4>: 'print'}, {<T.id: 1>: 1026, <T.label: 4>: 'edit'}, {<T.id: 1>: 1027, <T.label: 4>: 'located_in'}, {<T.id: 1>: 1022, <T.label: 4>: 'edit'}, {<T.id: 1>: 1023, <T.label: 4>: 'edit'}]
"""WTF is __?
"A GraphTraversal<S,E> is spawned from a GraphTraversalSource. It can also be spawned anonymously (i.e. empty) via __."
- From the documentation
Basically, to pass a traversal into a function, add a starting point for the traversal. Use __ and build a traversal on it. usually, the function that gets this traversal will connect it to the previous traversal, as depicted in the following:
g.V('id_x').outE().filter(__.outE().hasId(2))`
# → will refer as `__` as starting point for the filtering,
# so the starting point will be
g.V('id_x').outE()Note: sometimes you won’t see the actual use of __, but take our word for it: you will use it a lot. When importing outE() functions (for example) that can act without being on top of a traversal, it actually aliases for __.outE().
For more details about __ see https://tinkerpop.apache.org/docs/3.5.0-SNAPSHOT/upgrade/#_anonymous_child_traversals
When to use it?
Whenever you need to pass a traversal to a function, you can use explicit __ and add anything to it, or import it from gremlin_python.process.graph_traversal. For example, you can import from graph_traversal.hasId() that is equal to __.hasId().
In version 3.5 of Gremlin-Python and so forth, it is mandatory to use the __ instead of .g in the middle of queries.
Need to Use a Python Keyword as a Step Like min, id, or in?
Add an underscore "_" suffix
From the docs:
When Python reserved words and global functions cross paths with ordinary Gremlin steps and tokens, the conflicting Gremlin bits are suffixed with an underscore:
Steps - and_(), as_(), filter_(), from_(), id_(), is_(), in_(), max_(), min_(), not_(), or_(), range_(), sum_(), with_()
# Example
# g.V(4).in().toList() # will raise a `SyntaxError`
g.V(4).in_().toList()outE vs inV? Shouldn’t This Be the Other Way Around?
From the PRACTICAL GREMLIN Tutorial - https://kelvinlawrence.net/book/Gremlin-Graph-Guide.html
- out -> Outgoing adjacent vertices.
- in -> Incoming adjacent vertices.
- both -> Both incoming and outgoing adjacent vertices.
- outE -> Outgoing incident edges.
- inE -> Incoming incident edges.
- bothE -> Both outgoing and incoming incident edges.
- outV -> Outgoing vertex.
- inV -> Incoming vertex.
- otherV -> The vertex that was not the vertex we came from.
Debugging Tips
Here are some useful one-liners to copy and paste when you pause a Python debugger with a GraphTraversalSource object to play around with:
Unsure Whether Your Traversal is Working as Intended?
.path().next(). You can start from the first non-obvious call and work from there, adding one traversal step at a time, until you find the issue. Alternatively, binary search the step that’s misbehaving.
# Example:
g.V().out("edit").hasLabel("file").toList()
# [v[3], v[6]]To get to know the path the traversal went through, use:
g.V().out("edit").hasLabel("file").path().toList()
# [path[v[1], v[3]], path[v[2], v[6]]]
# Another example:
g.V().outE().inV().outE().inV().path().toList()
# [path[v[1], e[9][1-edit->2], v[2], e[12][2-print->5], v[5]],
# path[v[1], e[9][1-edit->2], v[2], e[13][2-edit->6], v[6]],
# path[v[1], e[10][1-edit->3], v[3], e[14][3-located_in->7], v[7]]]As you can see, the traversal went from v[1] and arrived at v[7] via v[3].
Traversal Profiling
To optimize traversals, see what the traversal is doing behind the scenes and how time-consuming every step is:
g.V().out("edit").out().profile().next()
"""
{'dur': 0.787151,
'metrics': [{'dur': 0.144021,
'counts': {'traverserCount': 7, 'elementCount': 7},
'name': 'TinkerGraphStep(vertex,[])',
'annotations': {'percentDur': 18.29648949185099},
'id': '6.0.0()'},
{'dur': 0.125388,
'counts': {'traverserCount': 3, 'elementCount': 3},
'name': 'VertexStep(OUT,[edit],vertex)',
'annotations': {'percentDur': 15.92934519552157},
'id': '1.0.0()'},
{'dur': 0.093565,
'counts': {'traverserCount': 3, 'elementCount': 3},
'name': 'NoOpBarrierStep(2500)',
'annotations': {'percentDur': 11.886537652877275},
'id': '5.0.0()'},
{'dur': 0.424177,
'counts': {'traverserCount': 3, 'elementCount': 3},
'name': 'VertexStep(OUT,vertex)',
'annotations': {'percentDur': 53.887627659750166},
'id': '2.0.0()'}]}
"""The total duration (0.78ms) is visible, and for each part of the query, you can see the traversal count and the time share (percentage) of each part. Note that the traversal starts from all of the 7 vertices, then goes down to the 3 out edit edges.
How to filter queries efficiently
There are many options to filter traversals, one option is to explicitly add a filter step.
With the filter command, you can filter a traversal by a query that consists of another traversal:
# filter vertices with at least 2 out edges
g.V().filter(__.outE().count().is_(P.gte(2))).toList()
# [v[1], v[2]]
# filter edges that connected to vertices with id in the list [4,7]
g.E().filter(__.inV().hasId(P.within([4, 7]))).toList()
# [e[11][1-view->4], e[14][3-located_in->7]]
g.V().filter(__.inE().hasLabel("print")).toList()
# [v[5]]According to the filter documentation: “Map the traverser to either true or false, where false will not pass the traverser to the next step.”
Another option is to use other filtering commands such as .or, .and, .has.
How Can I Save Values During Traversals?
If you want to traverse the graph and save a few values during the traversal, it can be useful to use a function that saves the values, as depicted in the following example:
from gremlin_python.process.graph_traversal import GraphTraversal
def save_value_from_traversal(query: GraphTraversal, property_name: str, saving_name: str) -> GraphTraversal:
"""Return query with saved value."""
return save_properties_from_traversal_impl(query, property_name, saving_name)
def save_id_from_traversal(query: GraphTraversal, saving_name: str) -> GraphTraversal:
"""Return query with saved id."""
return save_properties_from_traversal_impl(query, "id", saving_name)
def save_value_map_from_traversal(query: GraphTraversal, saving_name: str) -> GraphTraversal:
"""Return query with saved value map."""
return save_properties_from_traversal_impl(query, None, saving_name)
def save_label_from_traversal(query: GraphTraversal, saving_name: str) -> GraphTraversal:
"""Returns query with saved label."""
return save_properties_from_traversal_impl(query, "label", saving_name)
def save_properties_from_traversal_impl(query: GraphTraversal, property_name: Optional[str], value_saving_name: str) -> GraphTraversal:
"""
Return query with saved value. if the property name is 'id'/'label' will return the id/ label. if property is 'None' return all the properties.
:param query: Graph traversal
:param property_name: name of property we wish to save from the traversal - None for a valueMap
:param value_saving_name: the name to save the query result under
:return: a traversal that stores the property's value under value_step_label.
"""
origin_saving_name = "save_id_or_value_from_traversal_origin_query"
query = query.as_(origin_saving_name)
if property_name == "id":
query = query.id()
elif property_name == "label":
query = query.label()
elif not property_name:
query.valueMap()
else:
query = query.values(property_name)
query = query.as_(value_saving_name)
query = query.select(origin_saving_name)
return queryLet's look at an example and see how we can use the function that we defined:
# Without using the saving function:
q = g.V()
q = q.as_("first_vertex").label().as_("first_vertex_label").select("first_vertex") # can continue the traversla here
q.select("first_vertex_label").toSet()
# {'drive', 'file', 'user'}It is OK, but when saving many values, it’s better to avoid the mess:
# Using saving function
q = g.V()
q = save_label_from_traversal(q, "first_vertex_label")
q.select("first_vertex_label").toSet()
# {'drive', 'file', 'user'}Working with Neptune AWS DB
For small graphs, it is useful to develop on a local machine, but to scale up you will need a graph DB such as Neptune. Let’s go over some practical tips about it.
Timeout
It is common to encounter timeout problems when interacting with full-scale graphs, especially if you intend to run complex queries on them. This can be solved quickly by applying:
from datetime import timedelta
evaluation_timeout_milliseconds = timedelta(hours=1).total_seconds() * 1000
g = g.with_("evaluationTimeout", evaluation_timeout_milliseconds)Neptune Idle Time Problem
After some no-operation time (15 min), Neptune will close the connection. To avoid this, renew the connection after any long idle time/calculations. To control the connection and use many useful functions with Neptune, see gremlin_utils.py.
Neptune Error Handling
If your queries do not return or return with some errors (not enough RAM/CPU power to complete the query), try to run Neptune on a larger machine. You may still get internal errors, no reply, and timeout errors, but nothing related to the machine’s capacity.
Data-Driven Optimization
Traversal Starting Point
Sometimes, you have a traversal between two types of entities in the graph, and you can choose your starting entity. It is better to start from the entity that has fewer occurences in the graph.
g.V(1).repeat(__.addV("user")).times(1000).next() # adding 1000 users
v[1014]
g.V(1).repeat(__.addV("user")).times(1000).next() # adding 1000 users.V().label().groupCount().next()
{'drive': 1, 'user': 1002, 'file': 4}
%%timeit
g.V().hasLabel("drive").in_().in_().hasLabel("user").toList()
582 µs ± 39.5 µs per loop (mean ± std. dev. of 7 runs, 1,000 loops each)
%%timeit
g.V().hasLabel("user").out().out().hasLabel("drive").toList()
598 µs ± 16.2 µs per loop (mean ± std. dev. of 7 runs, 1,000 loops each)When the graph is fully loaded, it can vary significantly. You are encouraged to run these traversals with .profile() and see what happens.
Skip Stopping on Edges, Use out() Rather Than outE().inV()
The following query is the same as the above, but we add a tiny stop on the edge, this query will be slower than the previous one:
%%timeit
# will be slower than the previous traversal
g.V().hasLabel("user").outE().inV().outE().inV().hasLabel("drive").toList()
622 µs ± 12 µs per loop (mean ± std. dev. of 7 runs, 1,000 loops each)Gremlin Visualization
It will become super important for you to be able to dynamically navigate through the graph, so you can dive into highly dense areas or zoom out to see the full picture.
One option to do so is with the following library: https://github.com/prabushitha/gremlin-visualizer. You can download a docker from there and quickly visualize your queries:
docker run --rm -d -p 3000:3000 -p 3001:3001 --name=gremlin-visualizer --network=host prabushitha/gremlin-visualizer:latest
from IPython.display import IFrame
IFrame("http://localhost:3000", 800, 600)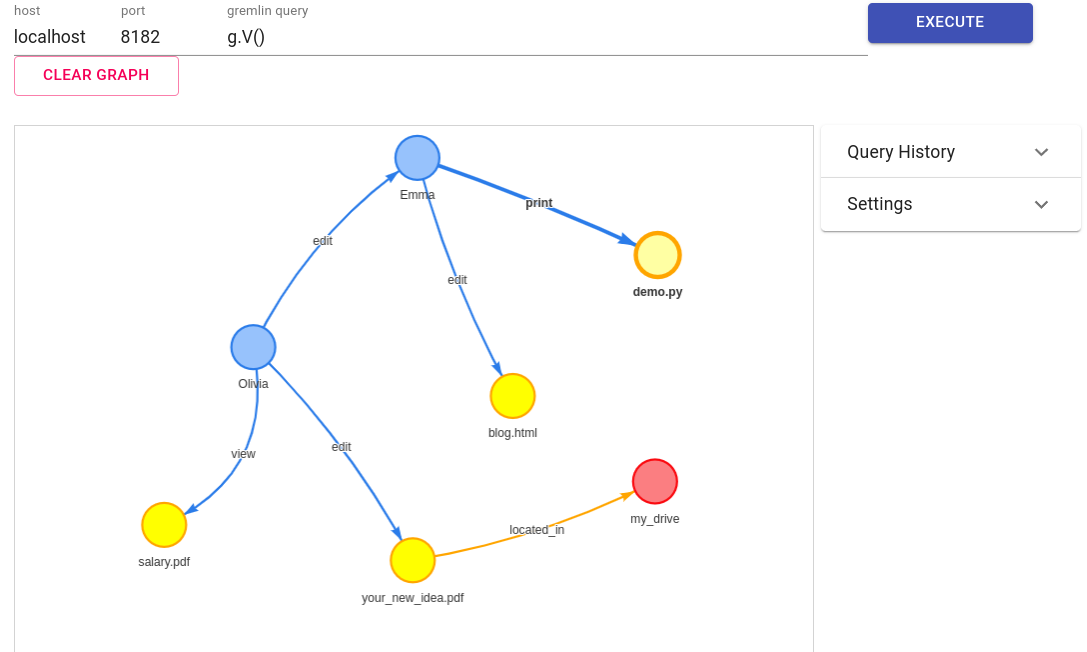
Fast Transformation to Networkx
You might want to run some algorithms with a graph library such as NetworkX, specifically, to look at the graph relations (and less on properties). It can also scale up (you can even hold a graph with tens of millions of edges and vertices on your local machine). There are some benefits in using such a library for developing algorithms:
- Advanced built-in algorithms
- Significantly faster access time
- Many web sources about algorithms implementation in NetworkX
There are many ways to export your data to NetworkX, but there are also many configurations that you may want to perform, so you might want to know how to do it yourself.
Here’s an example of how to rapidly export our graph to MultiDiGraph NetworkX, without properties:
import networkx as nx
vertices_list = [(v.id, {"label": v.label}) for v in g.V().toList()]
edge_list = [(edge.outV.id, edge.inV.id, {"label": edge.label}) for edge in g.E().toList()]
NX_graph = nx.MultiDiGraph()
NX_graph.add_nodes_from(vertices_list)
NX_graph.add_edges_from(edge_list)
print(f"{NX_graph.number_of_edges()=}")
print(f"{NX_graph.number_of_nodes()=}")
NX_graph.number_of_edges()=6
NX_graph.number_of_nodes()=7Drawing a Dynamic Graph with ipycytoscape
https://github.com/cytoscape/ipycytoscape
import ipycytoscape
[137]:
graph_draw = ipycytoscape.CytoscapeWidget()
graph_draw.graph.add_graph_from_networkx(NX_graph, directed=True)
graph_draw.set_style(
[
{
"selector": "any",
"style": {
"font-family": "helvetica",
"font-size": "10px",
},
},
{"selector": "any", "style": {"label": "data(label)"}},
]
)
display(graph_draw)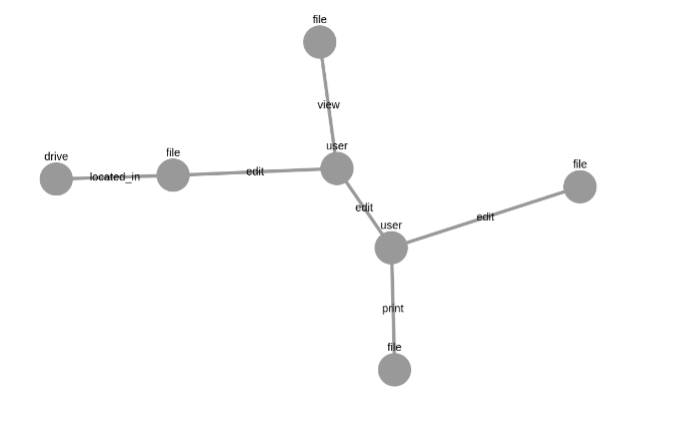
More Useful Links:
TinkerPop Documentation 👈 Official docs
PRACTICAL GREMLIN: An Apache TinkerPop Tutorial 👈 This is really good
